Admission CTAs
Mason scientists in I-SPY 2 trial discover first ever signatures of global resistance and response to prioritize breast cancer treatment selections, including in triple negative cases
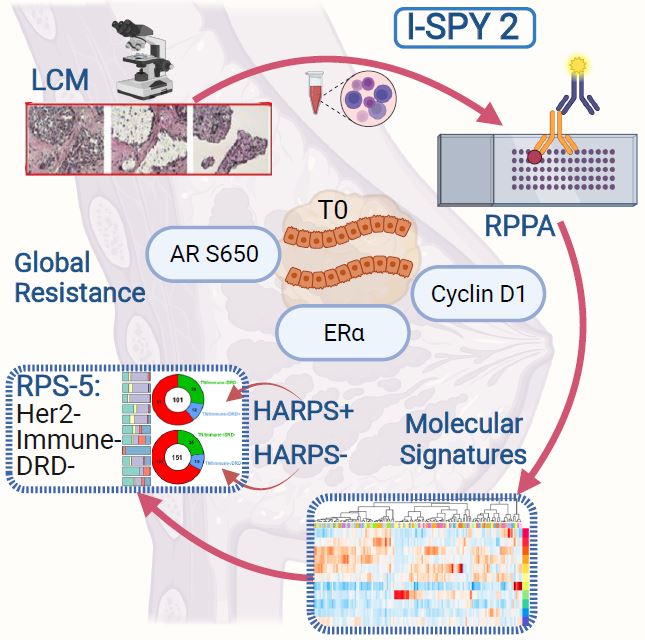
The proprietary LCM-RPPA protein activation mapping technique of breast tumors identifies three biomarkers associated with global resistance and response, several new drug targets and predictive signature that may help prioritize future treatment selection.
A team of George Mason University molecular biologists at its Center for Applied Proteomics and Molecular Medicine (CAPMM) has identified novel proteomic based signatures of global resistance for certain breast cancer patients and specific therapeutic strategies needed to overcome resistance. The work recently described in Cell Reports Medicine identifies new drug targets activated in human breast cancer, and described a new HER2-EGFR protein activation/phosphorylation signature called HARPS that could provide for better outcomes for patients with triple-negative breast (TNBC) cancer disease that previously had no specific precision therapy strategy identified.
Mason scientists Rosa Isela Gallagher, Julia Wulfkuhle, and Emanuel Petricoin, used their laser capture microdissection (LCM) and reverse phase protein array (RPPA) technology to map the protein drug target activation architecture of more than 700 breast tumors from the I-SPY 2 TRIAL, collaborating closely with genomics and bioinformatic scientists within the Departments of Laboratory Medicine and Surgery at University of California, San Francisco, and members of the I-SPY 2 TRIAL. This correlative study involved women with high-risk stage II and III early breast cancer who were enrolled in the first eight experimental arms of I-SPY 2 plus concurrent controls.

“This effort, the largest proteomic analysis of clinical tumor epithelium samples of any cancer type ever, is a culmination of a decade of work, in collaboration with dozens of clinicians and researchers in the I-SPY 2 TRIAL,” shared Emanuel Petricoin, co-director of Mason’s Center for Applied Proteomics and Molecular Medicine. “The team used the LCM-RPPA platform to map the protein drug target activation landscape of breast tumors prior to the patients receiving any therapy and then identified specific proteins that predicted response and resistance. No other precision oncology trial in the world is doing this,” said Petricoin, a Distinguished University Professor in the Mason Science School of Systems Biology.
The I-SPY 2 TRIAL, among the most widely known personalized medicine trials in the world today, pioneered the use of adaptive design strategies for rapid identification of targeted therapies that could benefit women with Stage II-III breast cancer. Over 2,750 women at 40 clinical sites around the US have enrolled in I-SPY 2 since its inception.
Researchers identified three specific proteins activated and expressed in human breast cancer that did not respond to any treatment. “The identified biomarkers associated with global resistance are cyclin D1, estrogen receptor alpha, and phosphorylated androgen receptor,” Mason Science Senior Research Scientist, Isela Gallagher said. “Because we found that global resistance to all drugs tested in the study was based on these 3 proteins being elevated and/or activated, we can target or turn off these proteins directly which will help the I-SPY2 TRIAL clinicians and their pharmaceutical partners prioritize new therapeutic strategies that target these proteins,” said Gallagher.
The team found that 11 specific protein activation-based signatures defined the breast cancers evaluated in the study, and identified specific precision therapeutics that would be optimized for best response. The CAPMM team also identified a novel signature called HARPS that can be used to stratify TNBC, the hardest to treat breast cancer subtype into HARPS+ who would best respond to anti-HER2 therapies or HARPS- who are predicted to best respond to immunotherapy. “The use of HARPS in TNBC could result in ~80% response rate compared to the current 30-40% response rate,” explained Mason Science Research Professor, Julia Wulfkuhle. “We hope that ongoing and planned rigorous clinical validation of HARPS as well as the numerous predictive and prognostic signatures we describe in our paper will allow the eventual use of these candidate biomarkers in the clinical management of breast cancer patients”, Wulfkuhle said.
“The CAPMM team at George Mason University has served as essentially the I-SPY protein biomarker engine since its inception,” shared Laura Esserman, I-SPY 2 TRIAL primary investigator and Professor of Surgery and Radiology at UCSF. “This novel approach gives our trials a powerful multi-omic molecular angle that is really unique in the precision oncology ecosystem,” explained Esserman, the Director of the UCSF Carol Franc Buck Breast Care Center.
“More importantly, the results give us clear targets that we can use to identify promising agents and test them in the I SPY 2.2 TRIAL- now that is bench to bedside translational research!” Esserman enthused.
“We are extremely inspired by the work being done by the team at George Mason University and are proud to be able to commercialize these efforts,” shared Faith Zaslavsky, President and CEO of CAPMM partner, Theralink Technologies. “Breakthroughs like this will save lives and dramatically reduce the cost of unnecessary and ineffective treatments.”
This research effort was supported by funding from many partners including the Quantum Leap Healthcare Collaborative, various National Institute of Health grants, the Gateway for Cancer research grants, The Atwater Trust and the SideOut Foundation. Further information and requests for resources or data should be directed to and will be fulfilled by Rosa I. Gallagher (rgallag3@gmu.edu).